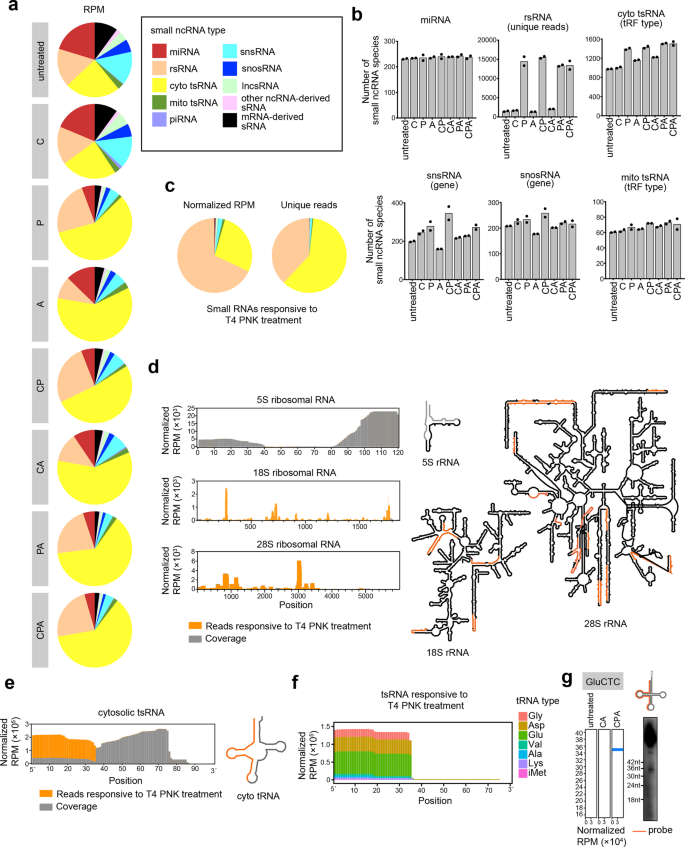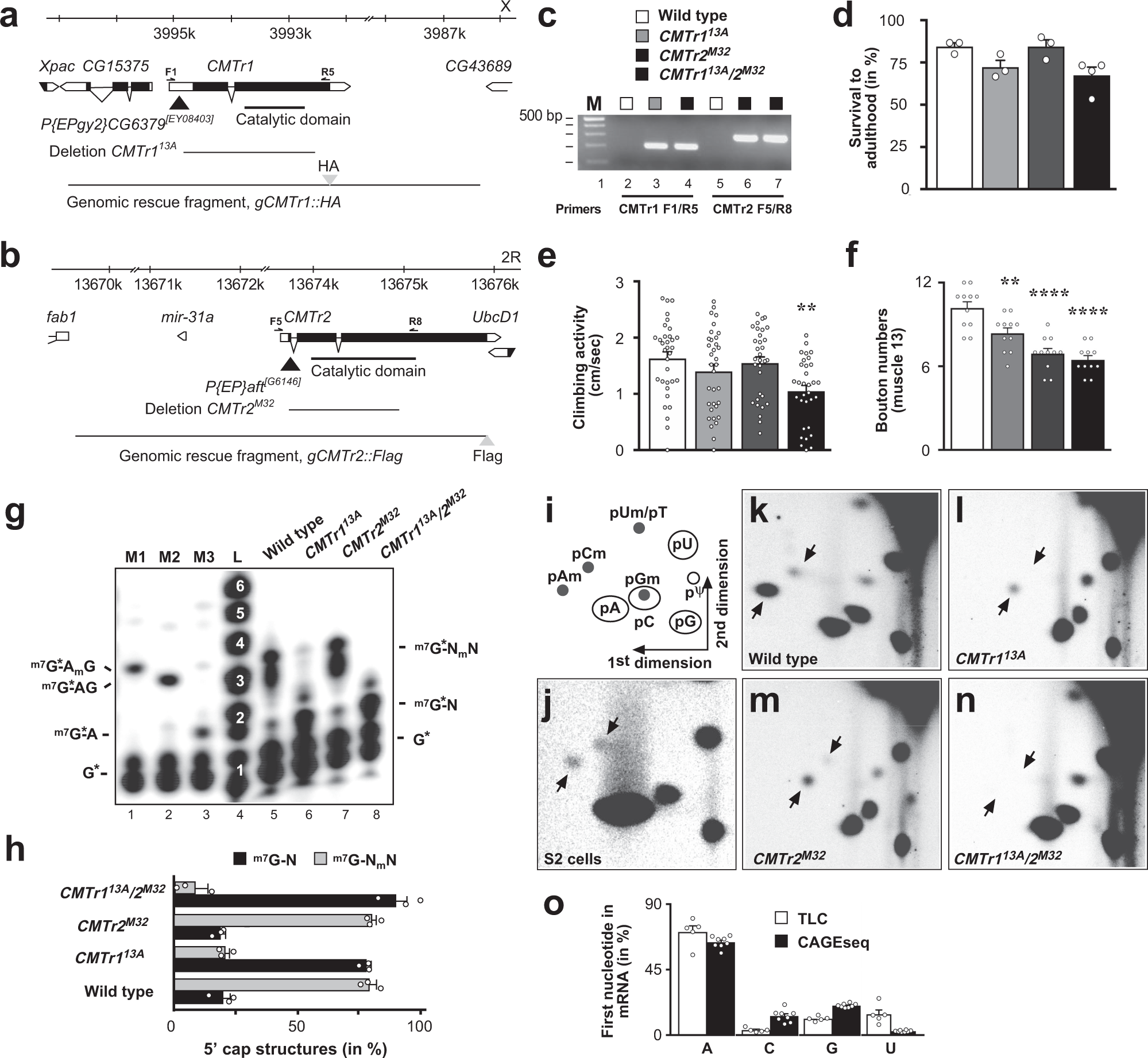
CMTr cap-adjacent 2′-O-ribose mRNA methyltransferases are required for reward learning and mRNA localization to synapses | Nature Communications

Thin layer chromatography reveals cap structures on sRNAs at all length... | Download Scientific Diagram

Messenger RNA cap methylation by PCIF1 attenuates the interferon-β induced antiviral state | bioRxiv
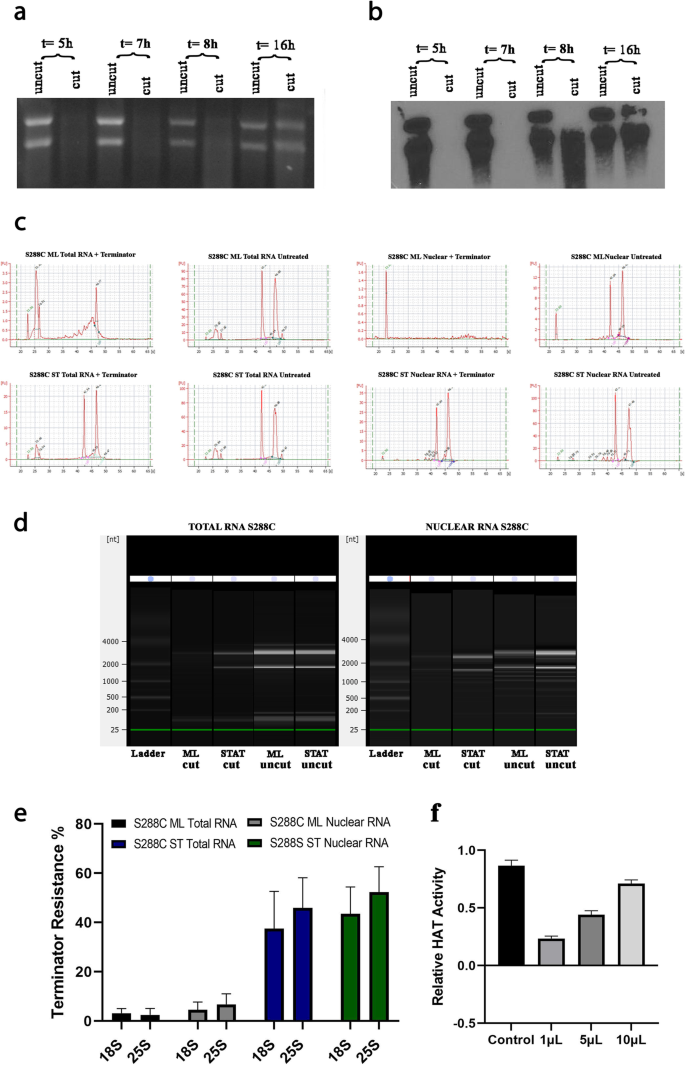
RNAP II produces capped 18S and 25S ribosomal RNAs resistant to 5′-monophosphate dependent processive 5′ to 3′ exonuclease in polymerase switched Saccharomyces cerevisiae | BMC Molecular and Cell Biology | Full Text
RNAP II produces capped 18S and 25S ribosomal RNAs resistant to 5'-monophosphate dependent processive 5'→3' exonuclease
CapClip Pyrophosphatase 5 U/µl | Phosphatasen und Kinasen | Enzyme | Biochemikalien | Biozym Scientific GmbH
Genome-wide quantification of 5'-phosphorylated mRNA degradation intermediates for analysis of ribosome dynamics - Document - Gale OneFile: Health and Medicine
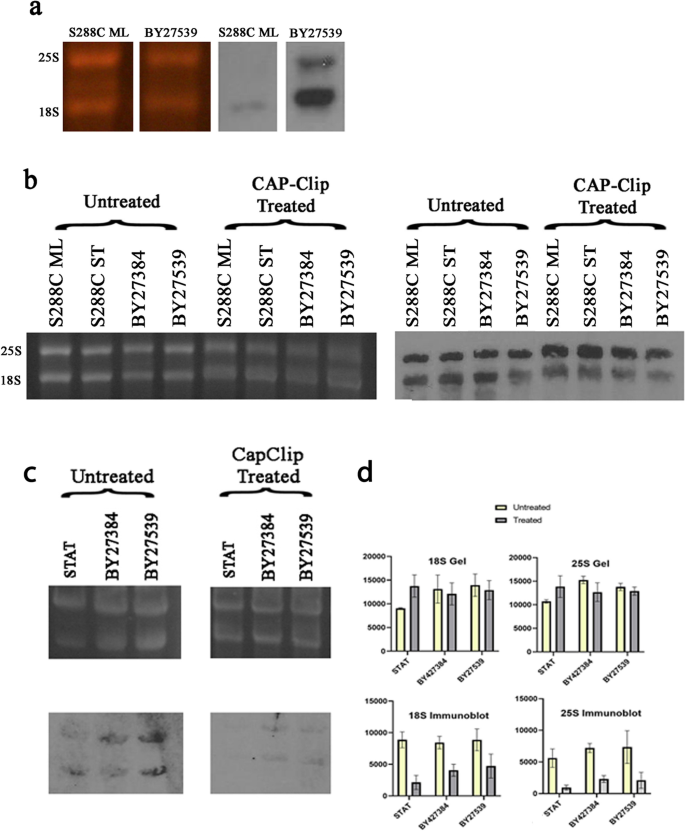
RNAP II produces capped 18S and 25S ribosomal RNAs resistant to 5′-monophosphate dependent processive 5′ to 3′ exonuclease in polymerase switched Saccharomyces cerevisiae | BMC Molecular and Cell Biology | Full Text

Methylation of viral mRNA cap structures by PCIF1 attenuates the antiviral activity of interferon-β | PNAS
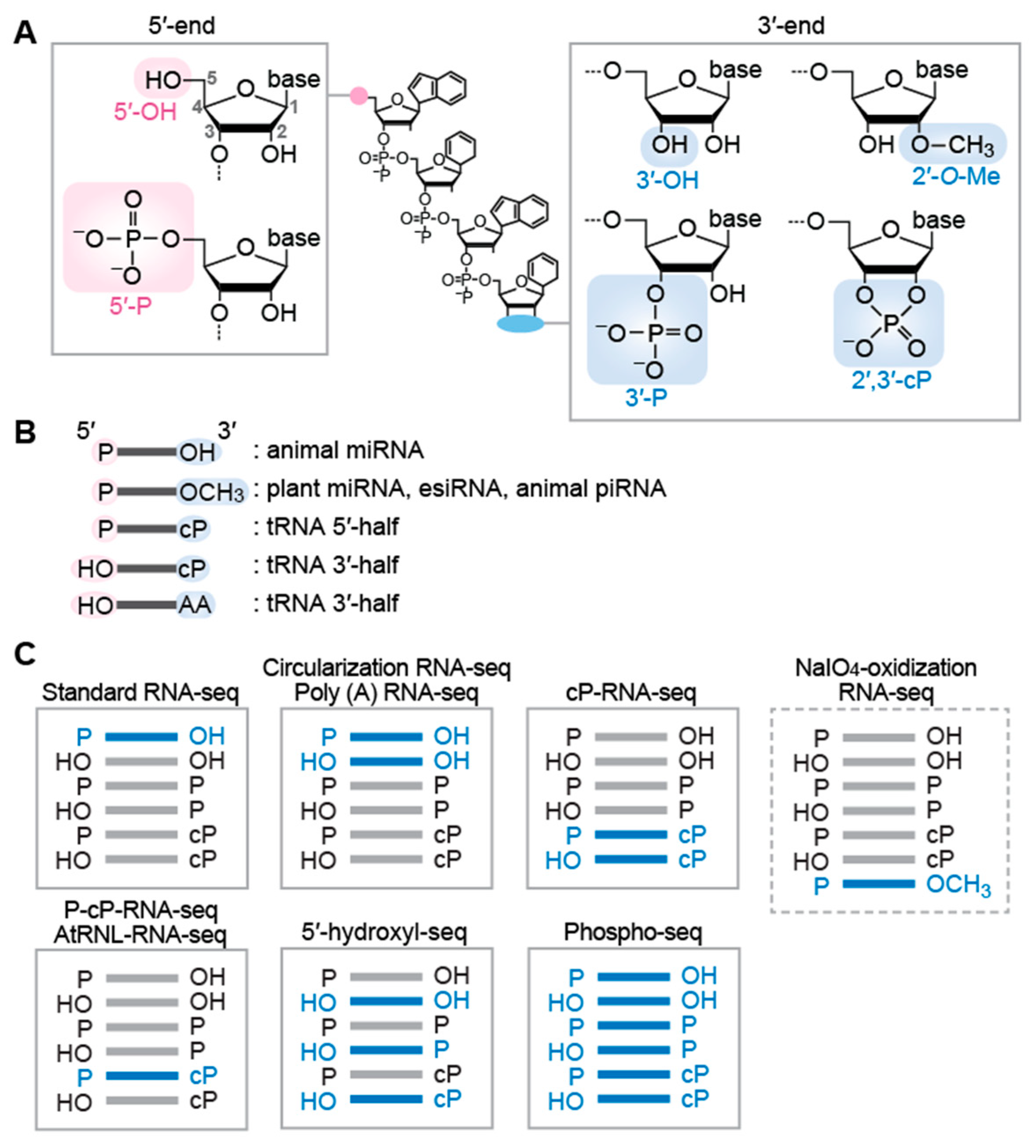
Biomolecules | Free Full-Text | Making Invisible RNA Visible: Discriminative Sequencing Methods for RNA Molecules with Specific Terminal Formations

Thin layer chromatography reveals cap structures on sRNAs at all length... | Download Scientific Diagram

Divergence of Structure and Function in the Haloacid Dehalogenase Enzyme Superfamily: Bacteroides thetaiotaomicron BT2127 Is an Inorganic Pyrophosphatase | Biochemistry

Structure and Sequence Requirements for RNA Capping at the Venezuelan Equine Encephalitis Virus RNA 5′ End | Journal of Virology

Structure and Sequence Requirements for RNA Capping at the Venezuelan Equine Encephalitis Virus RNA 5′ End | Journal of Virology